Waters Lab at Providence College
Our mission in the lab is to promote discovery and growth as comparative physiologists, complexity scientists, and stewards of nature and biodiversity.
We study ants and other insects, exploring their natural history, collective behavior, and metabolic physiology.
We strive to be an interdisciplinary and inclusive group and believe that all are welcome.
©2024 JSW | Template by Bootstrapious.com & ported to Hugo by Kishan B
Allometry with R and ggplot2
This web page was created based on using the blogdown and Hugo packages to publish an R Markdown document. Markdown is a simple formatting syntax for authoring HTML, PDF, and MS Word documents. For more details on using R Markdown see http://rmarkdown.rstudio.com.
In the text below, you should be able to copy all of the code in the gray code chunks and paste it into a script file to replicate the results and figures in R on your own computer.
Making a figure demonstrating the metabolic allometry of social insect colonies
# loading libraries
library(ggplot2)
library(scales)
# Importing the raw data
# Doing this with dput() on a data set I previously assembled instead of making you import a separate data file. For simplicity, so you can execute this whole page of code without needing anything else.
scaling.data <- structure(list(Mass_g = c(1.4, 1.7002, 1.8, 1.9434, 2.1375, 2.1416,
2.499, 2.5802, 2.82, 3.14, 3.15, 3.2887, 3.9303, 3.9844, 4.5304,
6.22, 11.15, 0.5831, 2.1449, 2.3775, 2.4805, 2.772, 2.83, 3.2861,
3.6833, 4.12, 4.2299, 4.4661, 7.23, 8.05, 11.12, 14.07, 15.994,
16.52, 23, 35, 0.026297706, 0.040436744, 0.044237498, 0.045942821,
0.062768431, 0.07691959, 0.065188108, 0.065188108, 0.066432704,
0.095607687, 0.102634562, 0.109658189, 0.104100736, 0.104100736,
0.102150441, 0.119965244, 0.149818237, 0.183594797, 0.169415356,
0.297376027, 0.187100058, 0.211573861, 0.316227766, 0.524460187,
0.615924842, 0.502611051, 0.502611051, 0.455104241, 0.455104241,
2.2228, 1.0851, 0.311, 1.0518, 1.947, 1.339, 1.7183, 1.8048,
1.8124, 0.3163, 0.4948, 1.0259, 0.4044, 0.199643, 0.129236, 0.171042,
0.073814, 0.216383228, 0.428173, 0.0735, 0.02291, 0.306489, 0.02648,
0.035914, 0.215908, 0.5765, 0.37279, 0.28873, 0.11961, 0.055239,
4.55, 9, 9.5, 20, 28, 46.5, 55.8, 56.7, 85.1, 93.3, 164, 170,
179, 195, 416, 438, 603, 839, 1150), MetRate_uW = c(11725, 10887.5,
11334.1666666667, 12221.9166666667, 14684.1666666667, 10775.8333333333,
17531.6666666667, 13902.5, 17587.5, 14963.3333333333, 17140.8333333333,
18648.3333333333, 23896.6666666667, 24120, 25236.6666666667,
28084.1666666667, 35621.6666666667, 6700, 13232.5, 14740, 19876.6666666667,
21719.1666666667, 26409.1666666667, 24734.1666666667, 27805,
23561.6666666667, 27079.1666666667, 30875.8333333333, 28810,
31545.8333333333, 45895, 66218.3333333333, 53516.25, 57675.8333333333,
71020, 79283.3333333333, 33.7753585, 30.92522114, 27.53816388,
26.65809962, 32.69596607, 39.00034792, 42.59470698, 45.24307437,
62.31533437, 53.71694463, 53.71694463, 53.71694463, 60.88626912,
67.11765019, 90.74424905, 85.82973089, 87.43759511, 77.14186669,
100.9640395, 92.01623324, 149.0857861, 146.3442914, 166.6475603,
178.6593132, 201.5666172, 207.2570381, 238.2114803, 255.3814733,
360.0043728, 2416.22338, 1440.06502, 381.9876907, 1162.588857,
2473.06643, 1777.550277, 2571.259972, 1251.548476, 1988.6147,
684.5054588, 847.744214, 1094.032191, 720.4679038, 1275.378835,
862.7678496, 1036.910662, 664.1199683, 1148.200077, 5802.41761,
426.9794844, 411.6824213, 2421.204683, 556.5702842, 501.2094544,
2049.271617, 4622.855531, 3281.145231, 2225.307599, 959.549531,
524.668946, 214411.1667, 368835, 341057.9167, 740350, 1009913.333,
1090425, 1000075.5, 949725, 1192605.583, 1479427, 1263620, 1898333.333,
1828932.5, 2743650, 2369120, 3105785, 3501420, 3513312.5, 5168770.833
), Species = structure(c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 13L, 13L, 13L, 13L,
13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L,
13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 12L,
12L, 12L, 12L, 12L, 12L, 12L, 12L, 12L, 12L, 12L, 12L, 12L, 11L,
11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L,
11L, 11L, 11L, 14L, 14L, 14L, 14L, 14L, 14L, 14L, 14L, 14L, 14L,
14L, 14L, 14L, 14L, 14L, 14L, 14L, 14L, 14L), .Label = c("Camponotus rufipes",
"Odontomachus bauri", "Za", "Anoplolepis steinergroeveri", "Atta columbica",
"Camponotus fulvopilosus", "Camponotus maculatus", "Eciton hamatum",
"Formica rufa", "Messor pergandei", "Pheidole dentata", "Pogonomyrmex californicus",
"Temnothorax rugatulus", "Apis mellifera"), class = "factor"),
Unit = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L), .Label = c("Colony",
"Worker ant"), class = "factor"), Type = structure(c(2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L,
6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L,
6L, 6L, 6L, 6L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L,
8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L), .Label = c("Camponotus rufipes",
"Odontomachus bauri", "Za", "Pheidole dentata", "Pogonomyrmex californicus",
"Temnothorax rugatulus", "Worker ant", "Apis mellifera"), class = "factor")), .Names = c("Mass_g",
"MetRate_uW", "Species", "Unit", "Type"), row.names = c(1L, 2L,
3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 13L, 14L, 15L, 16L,
17L, 18L, 19L, 20L, 21L, 22L, 23L, 24L, 25L, 26L, 27L, 28L, 29L,
30L, 31L, 32L, 33L, 34L, 35L, 36L, 37L, 38L, 39L, 40L, 41L, 42L,
43L, 44L, 45L, 46L, 47L, 48L, 49L, 50L, 51L, 52L, 53L, 54L, 55L,
56L, 57L, 58L, 59L, 60L, 61L, 62L, 63L, 64L, 65L, 66L, 67L, 68L,
69L, 70L, 71L, 72L, 73L, 74L, 75L, 76L, 77L, 78L, 79L, 80L, 81L,
82L, 83L, 84L, 85L, 86L, 87L, 88L, 89L, 90L, 91L, 92L, 93L, 94L,
95L, 226L, 227L, 228L, 229L, 230L, 231L, 232L, 233L, 234L, 235L,
236L, 237L, 238L, 239L, 240L, 241L, 242L, 243L, 244L), class = "data.frame")
# Inspect data frame
head(scaling.data)
## Mass_g MetRate_uW Species Unit Type
## 1 1.4000 11725.00 Odontomachus bauri Colony Odontomachus bauri
## 2 1.7002 10887.50 Odontomachus bauri Colony Odontomachus bauri
## 3 1.8000 11334.17 Odontomachus bauri Colony Odontomachus bauri
## 4 1.9434 12221.92 Odontomachus bauri Colony Odontomachus bauri
## 5 2.1375 14684.17 Odontomachus bauri Colony Odontomachus bauri
## 6 2.1416 10775.83 Odontomachus bauri Colony Odontomachus bauri
str(scaling.data)
## 'data.frame': 114 obs. of 5 variables:
## $ Mass_g : num 1.4 1.7 1.8 1.94 2.14 ...
## $ MetRate_uW: num 11725 10888 11334 12222 14684 ...
## $ Species : Factor w/ 14 levels "Camponotus rufipes",..: 2 2 2 2 2 2 2 2 2 2 ...
## $ Unit : Factor w/ 2 levels "Colony","Worker ant": 1 1 1 1 1 1 1 1 1 1 ...
## $ Type : Factor w/ 8 levels "Camponotus rufipes",..: 2 2 2 2 2 2 2 2 2 2 ...
# Plotting the raw data
ggplot(scaling.data, aes(x=Mass_g, y=MetRate_uW)) + geom_point()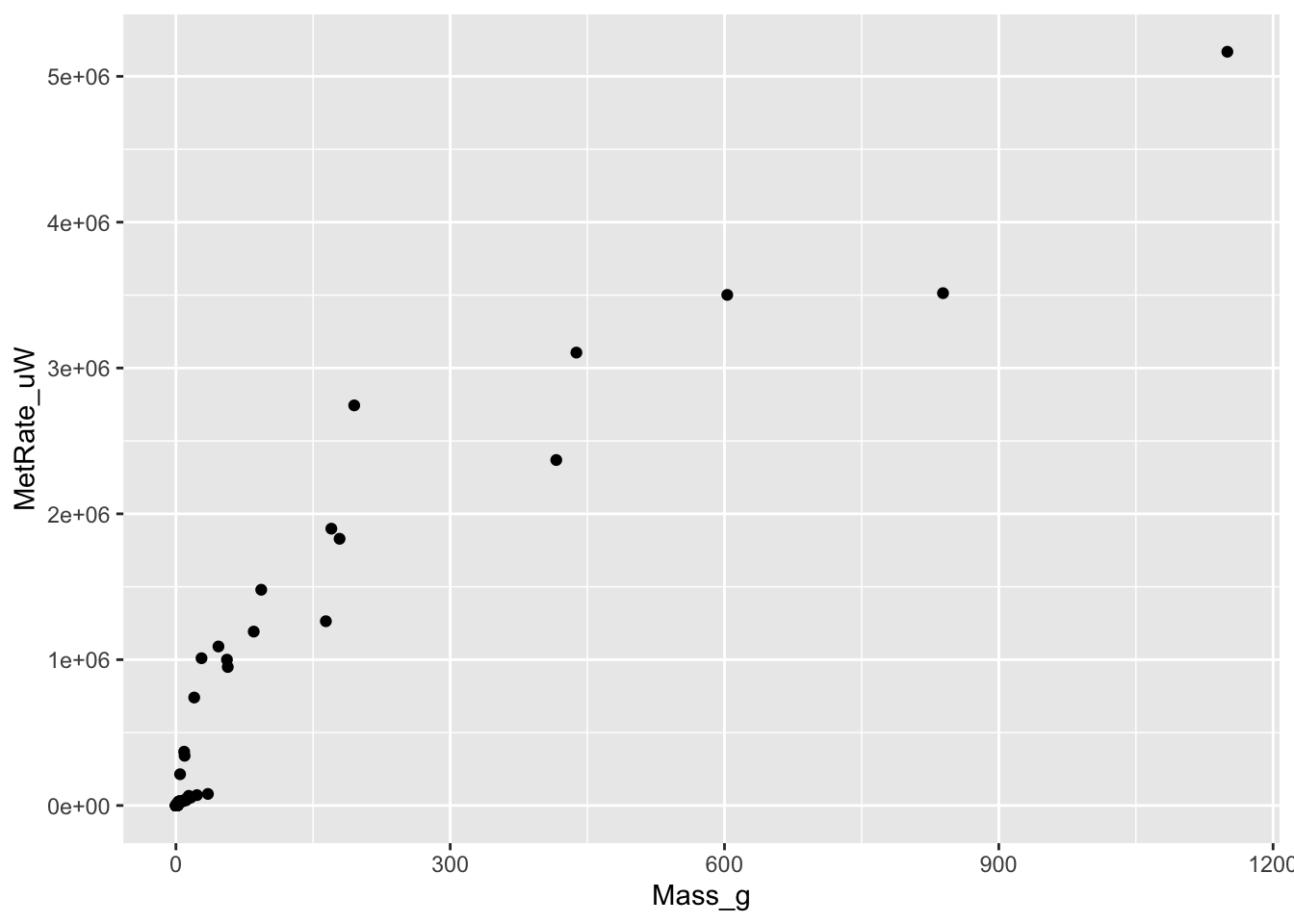
# Log-transforming the data on x- and y-axes
ggplot(scaling.data, aes(x=log10(Mass_g), y=log10(MetRate_uW))) + geom_point()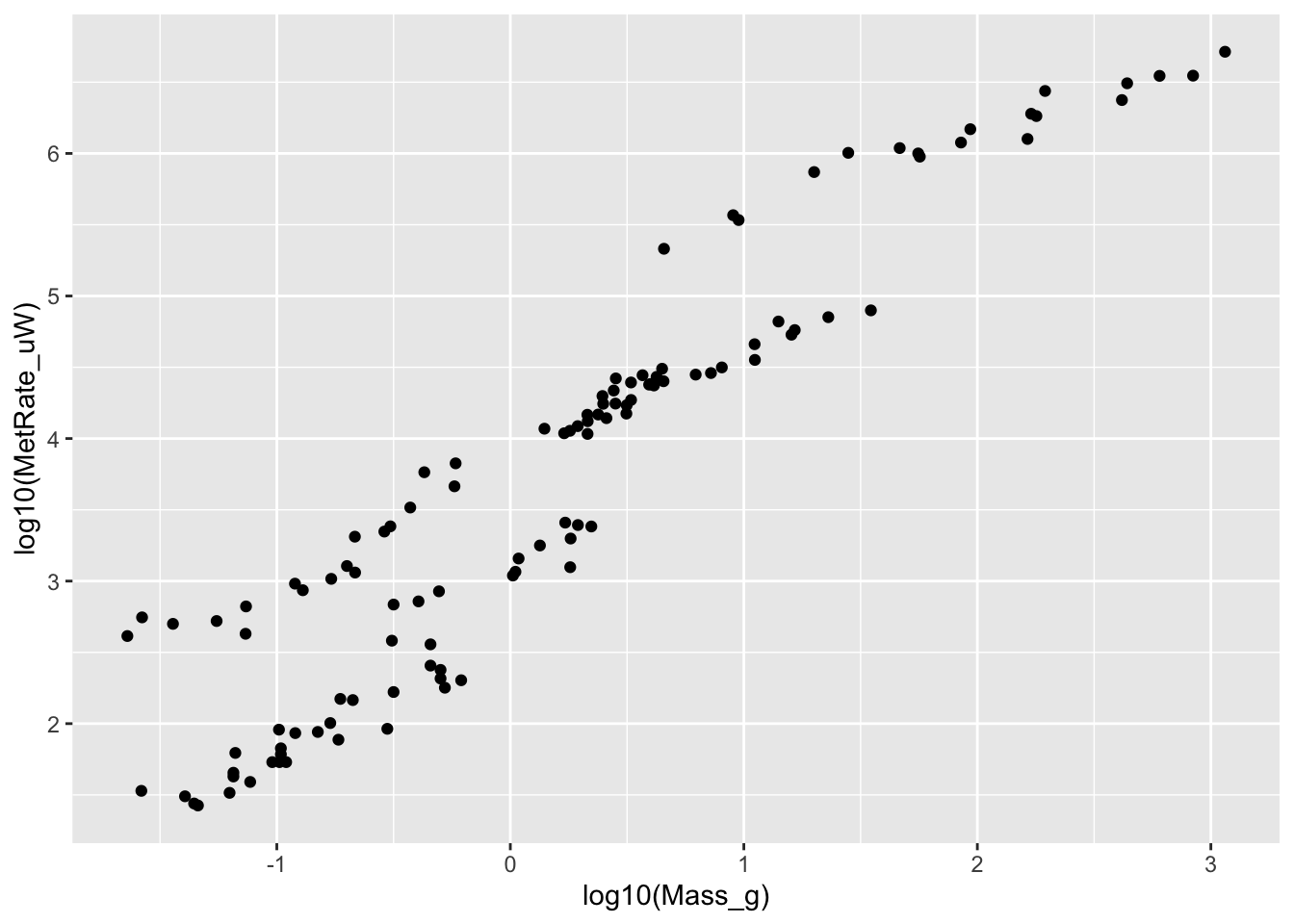
# Adding color codes and regression lines
ggplot(scaling.data, aes(x=log10(Mass_g), y=log10(MetRate_uW), color=Type)) + geom_point() + stat_smooth(method="lm")
## `geom_smooth()` using formula = 'y ~ x'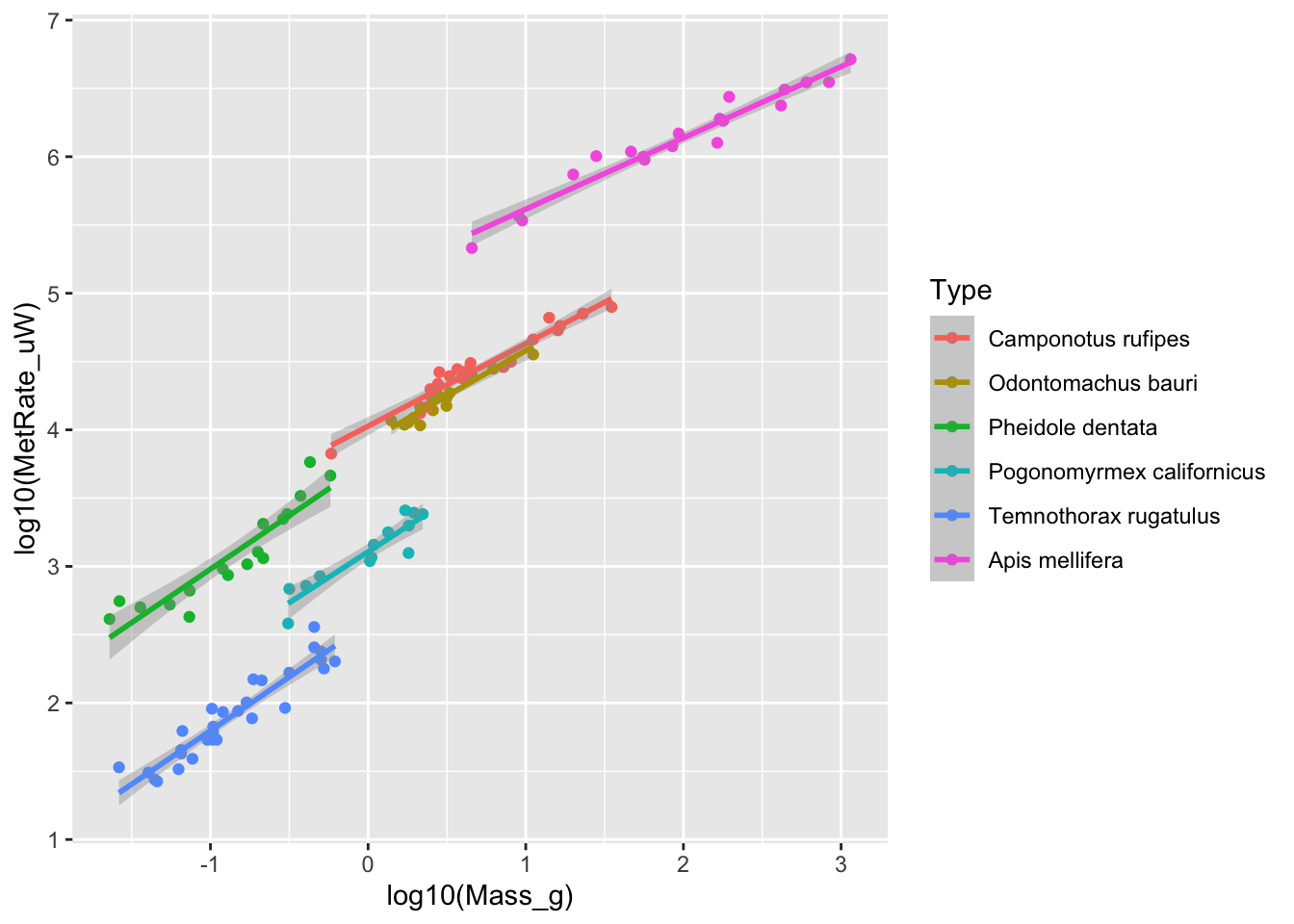
# Making it fancy by adding
# - log ticks on the axes
# - axes labels appropriate for log transformed data
# - simplifying the ggplot theme look
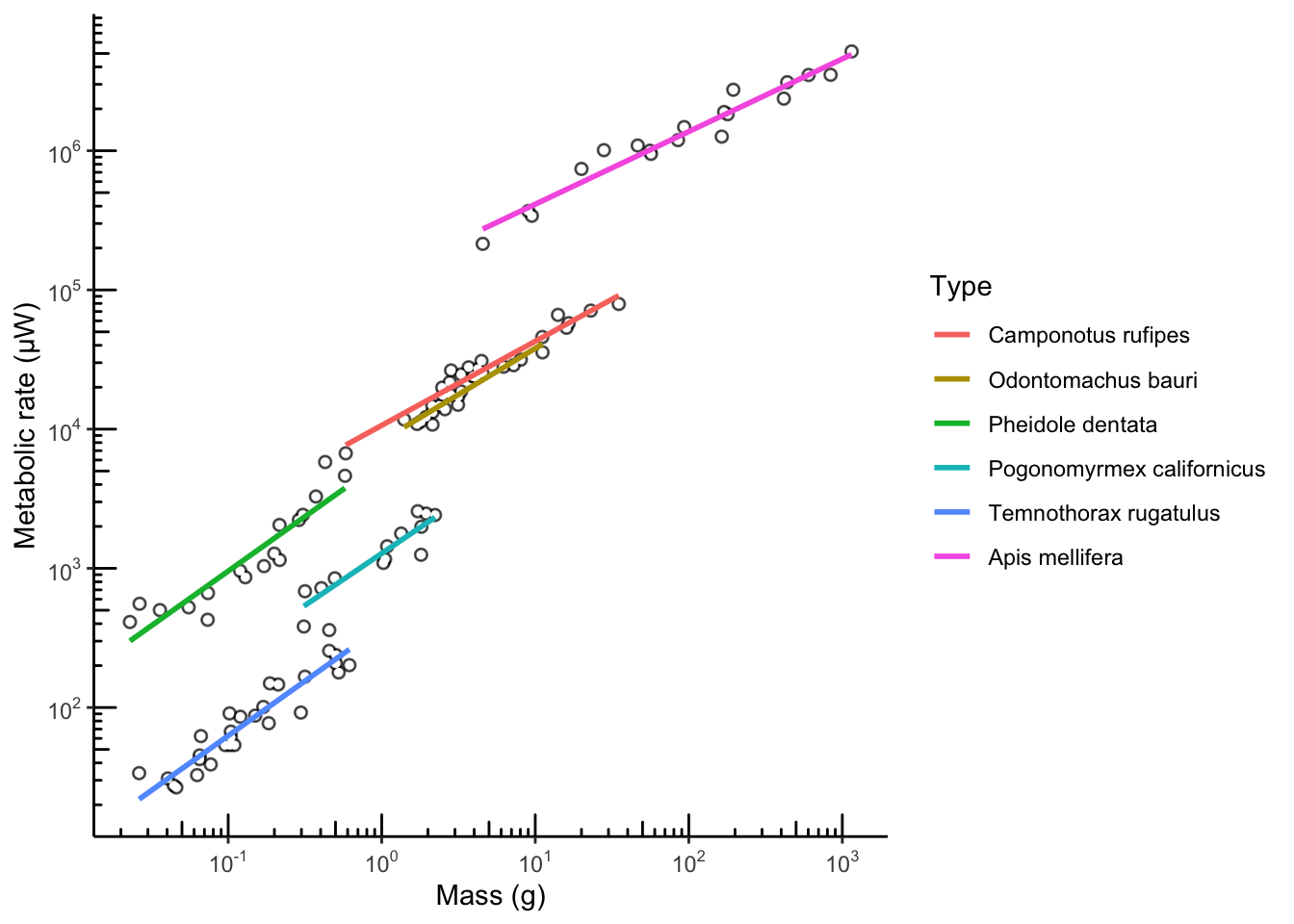
ggplot(scaling.data, aes(x=log10(Mass_g), y=log10(MetRate_uW), color=Type)) + geom_point(size=1, color="grey80") + annotate("point", x=log10(scaling.data$Mass_g[scaling.data$Unit == "Colony"]), y=log10(scaling.data$MetRate_uW[scaling.data$Unit == "Colony"]), color="black", size=2, alpha=0.6) + annotate("point", x=log10(scaling.data$Mass_g[scaling.data$Unit == "Colony"]), y=log10(scaling.data$MetRate_uW[scaling.data$Unit == "Colony"]), color="white", size=1) + geom_smooth(method="lm", se=F) + theme_bw() + theme(panel.grid.minor = element_blank()) + scale_x_continuous(name = "Mass (g)", labels = math_format(10^.x)) + scale_y_continuous(name = "Metabolic rate (µW)", labels = math_format(10^.x)) + annotation_logticks(sides="lb") + theme(panel.grid.major=element_blank(), panel.border=element_blank())+ theme(axis.line = element_line())
## `geom_smooth()` using formula = 'y ~ x'